BIOL 497/597 - Genomics & Bioinformatics
Syllabus
Sven Buerki - Boise State University
2026-01-07
1 Meeting Time and Place
- Day: Wednesday
- Time: 9:00 AM to 11:45 AM
- Location: Raptor Research Center Building, Rm 119
- Important Information: Students must bring their Bronco Card to access the building using key card readers located at the east, west, or north entrances. Because the Raptor Research Building is on the edge of campus, it is kept secure and requires card access for entry. Please carry your Bronco Card to every class session held in this building, as you will not be able to enter without it.
2 Instructor Information
- Name: Sven Buerki (he/his)
- Office location: Science building, office 228 (first floor)
- Office hours: By appointment
- Email address: svenbuerki@boisestate.edu
- Preferred way to contact me: By email or in class
3 Pre-requisite
BIOL 310: Genetics (3-0-3; F, S, SU).
A study of the principles of genetics as they relate to living organisms.
Prerequisites: BIOL 191–192 or BIOL 191 and BIOL 320.
Pre-/Corequisite: CHEM 301 or CHEM 307.
4 Welcome
Welcome to the course! I am looking forward to getting to know you this semester. To get started, please familiarize yourself with this syllabus and our website. I developed this course to provide a welcoming environment and effective learning experience for all students. If you encounter barriers in this course, please bring them to my attention so that I may work to address them, and reach out to me at any time if you have questions about course content or assignments.
5 Course Format
This is a face-to-face course that meets once per week. Each class session combines lecture, collaborative activities, and hands-on laboratory work designed to help you apply course concepts in real time with your peers.
A typical class meeting is structured as follows:
- An initial lecture component (ca. 50 minutes) introducing key concepts and methods
- A brief break and transition period
- A laboratory session (ca. 1 hour 40 minutes), which may include group activities, guided exercises, or computer-based data analysis
During the first several weeks of the semester, class time will emphasize lectures and structured group activities to build foundational knowledge and skills. As the semester progresses, the course will gradually transition to primarily computer-based laboratory work, with increased emphasis on independent and applied problem-solving.
To achieve the learning outcomes of the course, it is essential that you attend class regularly and actively engage with the material, your classmates, and the instructor.
You should expect to spend approximately 9 hours per week on this course, including both in-class instruction and out-of-class work such as readings, assignments, and project development.
6 Course Description, Goal, and Objectives
6.1 Goal
The primary goal of this course is to provide students with both theoretical and applied knowledge in genomics and bioinformatics required to sequence, assemble, and annotate eukaryotic genomes (Saitou, 2013; Satam et al., 2023), with particular emphasis on non-model organisms (Russell et al., 2017). The overall structure of the course and progression of topics are illustrated in Figure 6.1.
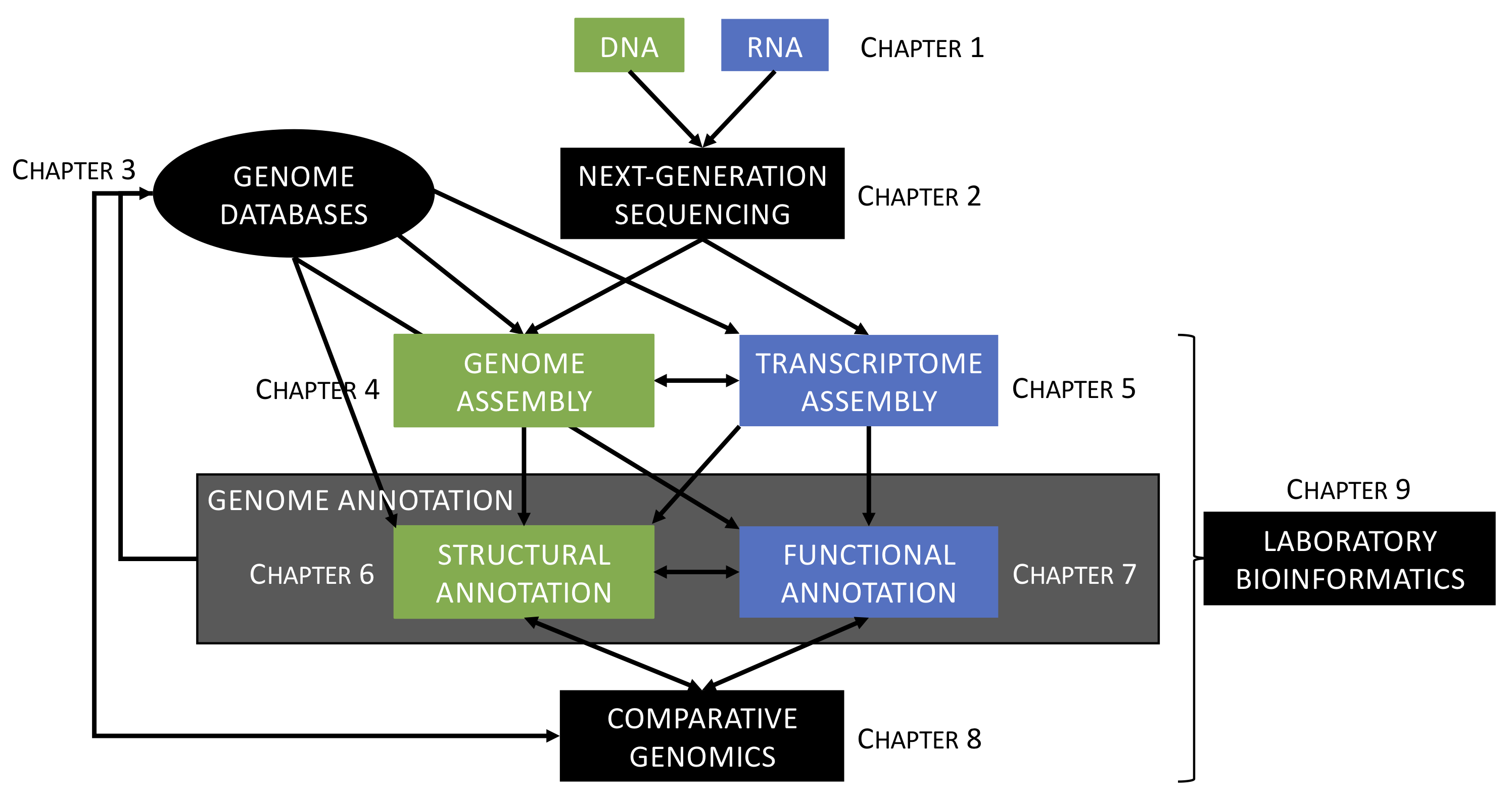
Figure 6.1: Overview of the structure of the course. See text for more details.
6.2 Objectives
To achieve this goal, the course is designed around three core objectives:
- Building strong genomics knowledge
- Developing practical bioinformatics skills
- Enhancing scientific dissemination abilities
Together, these objectives prepare students to engage with modern genomic research, from data generation and analysis to effective communication of results.
The first objective is to develop genomics knowledge through lectures that introduce foundational concepts in genome biology, sequencing technologies, and genome analysis, reinforced by applied examples that reflect real research workflows (Figure 6.1). To provide an authentic research experience, the course draws on examples and case studies from the instructor’s research program (Melton et al., 2021; e.g., Ellestad et al., 2022; Melton et al., 2022).
The second objective is to develop bioinformatics proficiency through hands-on use of a suite of unix-based open source software to perform genomic analyses, from raw sequence data to assembled and annotated genomes. Students will work through complete bioinformatics workflows commonly used to generate draft genome assemblies and annotations (Figure 6.2). In addition to learning how to execute bioinformatics programs, students will be taught how to install and manage software, gaining critical and transferable technical expertise. Reports and supporting documents provided by the instructor will give students a structured framework for analyzing and interpreting their own genomic datasets. Computational work will be conducted on computers running the Ubuntu Linux operating system, which is widely used in genomics research. This environment will allow students to become familiar with bash/shell, Python, and R, and will provide an on-ramp to using research computing clusters (e.g., the BSU Borah research cluster).
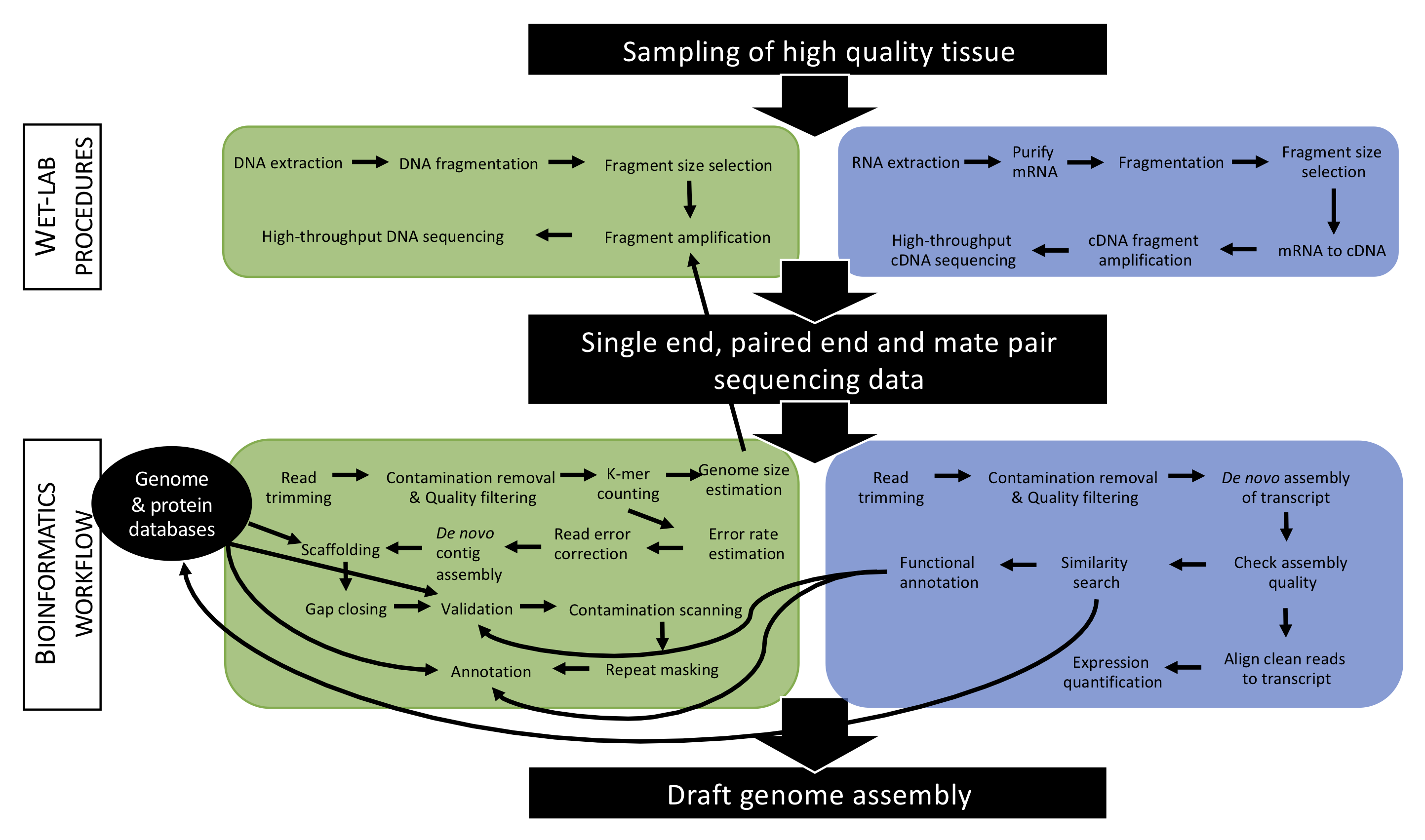
Figure 6.2: Overview of an example of an approach applied to produce a draft genome assembly. In this course, students will become accustomed with such approach and master some specific key steps.
The third objective is to foster scientific dissemination and critical evaluation by engaging students in writing, presenting, and discussing scientific papers and their own genomic analyses. Through these activities, students will develop the ability to clearly communicate results and to critically assess the methodological quality, rigor, and reproducibility of published genomic studies.
7 Course Content
Throughout the course, bioinformatics will be used as a central conduit for studying genomics, allowing students to explore genomic concepts through hands-on data analysis and computational workflows.
The course is subdivided into nine chapters distributed between lecture and laboratory sessions as follows (see Figure 6.1):
Lecture sessions:
- Chapter 1: DNA and RNA.
- Chapter 2: Next-generation sequencing.
- Chapter 3: Genome databases.
- Chapter 4: Genome assembly.
- Chapter 5: Transcriptome assembly.
- Chapter 6: Structural annotation.
- Chapter 7: Functional annotation.
- Chapter 8: Comparative genomics.
Laboratory sessions:
- Chapter 9: Applied bioinformatics.
8 Course Learning Outcomes
Below is a non-exhaustive list of learning outcomes associated with each chapter of the course (see Figure 6.1). Collectively, these outcomes align with three core learning outcomes that span the entire course and reflect its emphasis on genomics, bioinformatics, and scientific dissemination.
8.1 Core Learning Outcomes
By the end of this course, students will be able to:
Genomics: Explain key concepts in genome biology and critically evaluate genome sequencing, assembly, annotation, and comparative genomics strategies, with particular attention to eukaryotic and non-model organisms.
Bioinformatics: Apply bioinformatics tools and computational workflows to analyze genomic and transcriptomic data, including data quality assessment, assembly, annotation, and comparative analyses, using unix-based open-source software.
Scientific Dissemination: Effectively communicate genomic research through written reports and oral presentations, and critically evaluate published genomic studies for methodological rigor, data quality, and reproducibility.
8.2 Chapter-Specific Learning Outcomes
Note: Material associated with Chapters 5–8 (Transcriptome Assembly, Structural Annotation, Functional Annotation, and Comparative Genomics) will be studied primarily through Mini-Report 3 and the Group Lab Assignment (corresponding to Chapter 9). This approach is designed to help students engage with the material through authentic, evidence-based research, reinforcing learning by applying theoretical concepts to real genomic datasets.
8.2.1 Chapter 1: DNA & RNA
- Understand the differences between genetics and genomics.
- Understand the central dogma: DNA is transcribed into RNA, which is translated into protein.
- Appreciate the diversity of genome organization in organelles, prokaryotes, and eukaryotes.
- Understand how organelle genomes have contributed to the evolution of eukaryotic chromosomes and how this impacts genome assembly project design.
- Appreciate that eukaryotic genomes contain extensive and diverse repetitive regions.
- Appreciate the diversity of published genomes and variation in their levels of completeness.
- Understand the importance of computer science and bioinformatics in generating raw sequencing data and assembling genomes.
- Understand the major challenges associated with sequencing and assembling genomes.
8.2.2 Chapter 2: Next-Generation Sequencing
- Learn the terminology associated with next-generation sequencing (NGS) that underpins genome assembly and annotation.
- Understand the general principles of genome sequencing (wet-lab) and genome assembly (bioinformatics workflows) (see Figure 6.2).
- Be familiar with the major NGS platforms and their limitations (Satam et al., 2023):
- Illumina
- PacBio
- Oxford Nanopore
- Illumina
- Become proficient in handling NGS data outputs, particularly FASTA and FASTQ files.
- Learn how to assess nucleotide quality using Phred quality scores.
8.2.3 Chapter 3: Genome Databases
- Understand the role of computer science and bioinformatics in:
- Producing raw sequencing data,
- Creating molecular biology databases,
- Archiving and curating genomic data,
- Distributing data via the Internet,
- Developing tools for efficient data retrieval and mining.
- Gain knowledge of major molecular biology databases relevant to genome assembly and annotation, including:
- Nucleic acid sequence databases,
- Protein sequence databases,
- Gene ontology databases,
- Metabolic pathway databases,
- Specialized annotated genome portals.
- Learn protocols to query genomic information and remotely download data from GenBank.
8.2.4 Chapter 4: Genome Assembly
- Understand the key steps involved in producing a genome assembly and strategies to optimize assembly quality.
- Perform read-quality assessments, including:
- Read trimming,
- Contaminant screening,
- k-mer counting,
- Genome size estimation,
- Read error correction.
- Understand the use of de Bruijn graphs for de novo genome assembly.
- Gain an overview of commonly used genome assemblers and their specific applications.
- Understand the benefits of long-read sequencing for closing gaps and improving draft genome assemblies.
8.2.5 Chapter 5: Transcriptome Assembly
- Learn about the different types of RNA molecules in the cell.
- Understand the relationship between the transcriptome and proteome through the genetic code.
- Understand key steps involved in transcriptome assembly and strategies to optimize results, including comparisons between reference-based, de novo, and combined approaches.
- Gain an overview of commonly used transcriptome assemblers and their specific applications.
8.2.6 Chapter 6: Structural Annotation
- Understand what is meant by the term genome annotation.
- Understand key steps involved in structural genome annotation and best practices.
- Become familiar with major stages of the structural annotation process:
- Repeat identification,
- Evidence alignment,
- Ab initio gene prediction,
- Evidence-driven gene prediction,
- Overview of commonly used structural annotation software.
8.2.7 Chapter 7: Functional Annotation
- Understand the challenges associated with accurately assigning gene functions.
- Study and compare available pipelines for automated functional genome annotation.
- Review approaches used to assess annotation quality.
- Become familiar with commonly used functional annotation software.
- Gain an overview of procedures for submitting annotated genomes to public databases.
8.2.8 Chapter 8: Comparative Genomics
- Learn how to read, interpret, and present scientific papers reporting new genomic or transcriptomic data.
- Develop expertise through structured reading and discussion of scientific publications (e.g., Li et al., 2009; Ellestad et al., 2022).
8.2.9 Chapter 9: Applied Bioinformatics
In this chapter, students will use data produced by the instructor’s laboratory to learn practical bioinformatics protocols to:
- Analyze Sanger sequencing data,
- Conduct phylogenetic analyses,
- Perform quality control and cleaning of Illumina reads,
- Assemble draft genomes,
- Mine draft genome scaffolds for target genes,
- Validate gene mining by identifying open reading frames (ORFs), reconstructing protein sequences, and inferring gene phylogenetic trees,
- Annotate validated genes.
9 Course Schedule
The tentative schedule for this course is available here. The instructor wants to warn students that he might adjust the schedule to accommodate any needs. However, in case of changes, the instructor will make sure to contact enrolled students to keep them posted.
10 Course Resources
Teaching materials for this course will be shared with students via the dedicated course website, GitHub repository, and Google Drive.
10.2 Publications, Textbooks, and Online Resources
The reading materials for this course consist of a combination of scientific publications and textbook chapters (Dale et al., 2012; deSalle and Rosenfeld, 2013; Brown, 2017; Lesk, 2017). The instructor has copies of these textbooks, and students are welcome to consult them at any time.
A comprehensive list of references used in this course is provided here. In addition, there are numerous online resources dedicated to genomic and transcriptomic data. A selection is provided below:
Finally, the instructor has assembled a Lexicon containing definitions of key concepts used in this course. If you encounter a term that is not included in the Lexicon, please contact the instructor to have it added.
11 Bioinformatics Tools
Although we will be using software installed on the Linux computers, the instructor is advising students to install the following programs on their personal computers:
- The R statistical language that will allow you to gather data and analyze it.
- The Markdown and LaTeX markup languages that you can use to create documents (slideshows, articles, books, webpages) for presenting your findings.
- The knitr and rmarkdown packages for R and other tools, including command-line shell programs like GNU Make and Git version control, for dynamically tiding your data gathering, analysis, and presentation documents together so that they can be easily reproduced.
- RStudio, a program that brings all of these tools together in one place.
11.1 Installing Software
As shown above, R and RStudio are at the core of this course and will have to be installed on your computers. This can be easily done by downloading the software from the following websites:
The download pages for these software tools include detailed installation instructions; please refer to them for more information.
11.2 Installing Markup Software
If you are planning to create LaTeX documents, you will need to install a Tex distribution. Please refer to this website for more details: https://www.latex-project.org/get/
If you want to create Markdown documents you can separately install the rmarkdown package in R (see below for more details).
11.3 Installing R Packages
We will be using several R packages specifically designed to support reproducible research. Many of these packages are not included in the default R installation and must be installed separately.
To install the core packages used in this course, copy the following code and paste it into your R console:
install.packages(c("brew", "countrycode", "devtools", "dplyr", "ggplot2", "googleVis",
"knitr", "rmarkdown", "tidyr", "xtable"))Once you run this code, you may be prompted to select a CRAN “mirror” to download the packages from. Simply choose the mirror closest to your location.
It is also likely that we will need to install additional packages throughout the course. When that happens, you can install them using the same R function install.packages(), or through RStudio by selecting “Tools” → “Install Packages…”, entering the package name in the dialog box, and ensuring the “Install dependencies” option is checked.
11.4 RStudio Cheat Sheets
RStudio offers a collection of cheat sheets accessible from the “Help” menu by selecting “Cheatsheets.”
Five cheat sheets are especially relevant to chapters taught in this course:
- RStudio IDE : Cheat sheet
- Data Manipulation with dplyr, tidyr
- Data Visualization with ggplot2
- R Markdown Cheat Sheet
- R Markdown Reference Guide
These documents together with the material presented in course resources will provide the basis to design your bioinformatics tutorials.
11.5 R Tutorials
Please find below two documents providing a comprehensive introduction to R:
- R for beginners (a tutorial by Emmanuel Paradis): https://cran.r-project.org/doc/contrib/Paradis-rdebuts_en.pdf
- An introduction to R: https://cran.r-project.org/doc/manuals/r-release/R-intro.pdf
12 Assessments and Grading
Students will develop and demonstrate mastery of the course learning outcomes by producing written reports and delivering oral presentations, either individually or collaboratively in groups. Rather than relying on traditional exams, this course emphasizes hands-on application: you will build both theoretical genomics understanding and practical bioinformatics skills, and apply them directly to authentic research projects.
Graduate students: Graduate students enrolled in this course are held to a higher standard and are expected to take on leadership roles during group assignments. This includes guiding discussions, coordinating group efforts, and ensuring that analyses and reports meet the advanced expectations appropriate for graduate-level work.
12.1 Assessments
Students will be evaluated based on the following five mandatory assessments:
- Three individual mini-reports (2 × 25 points and 1 × 50 points; total: 100 points).
- One group lab report (150 points).
- One group lab presentation (50 points).
These assessments sum to a total of 300 points. Table 12.1 shows the grading scale used in this course.
12.2 Grading Policy
The grading scale for this course is in Table 12.1.
| Percentage | Grade |
|---|---|
| 100-98 | A+ |
| 97.9-93 | A |
| 92.9-90 | A- |
| 89.9-88 | B+ |
| 87.9-83 | B |
| 82.9-80 | B- |
| 79.9-78 | C+ |
| 77.9-73 | C |
| 72.9-70 | C- |
| 69.9-68 | D+ |
| 67.9-60 | D |
| 59.9-0 | F |
You can look at all of your scores by accessing Grades in the Canvas course menu.
12.3 Mini-Reports (TOTAL: 100 points)
To further develop expertise in genomics and bioinformatics, students will produce three mini-reports that align with the course’s core learning outcomes:
- Mini-Report 1: Sequencing technologies (25 points; related to Chapter 2)
- Genomics: Demonstrate understanding of genome sequencing principles and technologies.
- Bioinformatics: Explore how sequencing data are generated and processed.
- Scientific dissemination: Communicate technical information clearly in written form.
- Genomics: Demonstrate understanding of genome sequencing principles and technologies.
- Mini-Report 2: Molecular biology databases (25 points; related to Chapter 3)
- Genomics: Explain the role of databases in genome assembly and annotation.
- Bioinformatics: Apply protocols to query and retrieve genomic data.
- Scientific dissemination: Summarize and present information from multiple sources accurately.
- Genomics: Explain the role of databases in genome assembly and annotation.
- Mini-Report 3: DNA barcoding and phylogenetic analysis (50 points; related to Chapter 8)
- Genomics: Apply comparative genomics concepts to identify species and infer evolutionary relationships.
- Bioinformatics: Perform basic analyses, including sequence alignment and phylogenetic reconstruction.
- Scientific dissemination: Integrate methods and results into a coherent written report and justify the choice of comparative articles.
- Genomics: Apply comparative genomics concepts to identify species and infer evolutionary relationships.
Time will be allocated during class to work on these mini-reports; however, students are expected to complete them independently outside of class.
12.4 Group Lab Report (TOTAL: 150 points)
Students will work in groups to produce a report addressing the following research question:
What Aquaporin genes are encoded in the sagebrush genome (Artemisia tridentata) and what are their functions?
To gain further insights into Aquaporins, sagebrush, and the associated bioinformatic analyses, students will read the companion publication (Melton et al., 2021).
12.4.1 Associated Scripts
Students can access bash and R scripts associated with this lab report here.
12.4.2 Details on Reporting
Lab reports should be formal scientific documents, structured like manuscripts with the following sections: Introduction, Materials & Methods, Results, Discussion, and References.
- The instructor does not set a minimum word or page count but expects reports to be concise, clear, and free of redundancy.
- All evidence and documentation must be provided to allow an external reviewer to replicate analyses. Students are expected to include associated data and code to support reproducible science; guidance on these protocols will be provided during lab sessions.
- For group projects, each member must contribute equally. If participation is deemed insufficient, the instructor may assign individual grades.
12.4.3 Alignment with Course Learning Outcomes
- Genomics: Apply knowledge of genome structure and gene function to identify and interpret Aquaporin genes in a non-model eukaryotic genome.
- Bioinformatics: Use computational tools and scripts (
bashandR) to analyze sequence data, perform gene mining, and validate results.
- Scientific Dissemination: Communicate findings effectively through a structured written report, ensuring clarity, reproducibility, and proper scientific documentation.
12.5 Group Lab Presentation (TOTAL: 50 points)
Students will use their group lab reports to prepare 10-minute presentations, followed by 5 minutes for questions, scheduled during the final weeks of the semester.
- Each student is expected to actively participate in the presentation and contribute to answering questions from the audience.
- Presentations must be submitted in advance, either uploaded to the shared Google Drive folder or sent directly to the instructor.
12.5.1 Alignment with Course Learning Outcomes
- Genomics: Clearly explain the biological context, genome organization, and functional interpretation of Aquaporin genes in Artemisia tridentata.
- Bioinformatics: Summarize the computational analyses performed, including the use of
bashandRscripts, and demonstrate understanding of the bioinformatic workflow.
- Scientific Dissemination: Communicate research findings effectively in an oral format, respond to questions with clarity, and demonstrate the ability to convey complex genomic and bioinformatic concepts to peers.
12.6 Extra Credits
For students interested in earning extra credits, the instructor will provide two opportunities to earn a total of 30 extra credits. Students may allocate these extra credits to any of their assessments by sending an email to the instructor specifying the allocation.
These two non-mandatory assignments have strict deadlines (see Schedule). No extensions will be granted, and any work submitted after the deadline will not be considered. Students who do not submit by the deadline will be deemed not interested in earning the extra credits.
To support time management, the number of credits assigned to each question will be provided in advance. These opportunities allow students to improve their overall grade while gaining additional experience in genome assembly and annotation. As with other assignments, students may work in groups, but each student must submit their own individual assignment.
13 Expectation for Student Success
My goal is that every student is successful in this course, but I need your help to achieve that. In order to do your part to ensure your success in this course, please:
- Attend class. We will be using class time to practice and apply what we are learning, so it is important for your and your fellow students’ learning that you are present and participatory. You can miss two class periods without it affecting your grade. If you have to miss beyond two classes, please email me as soon as possible to discuss ways to help you participate in classroom activities asynchronously as appropriate.
- Ask questions. Learning is all about asking questions, so always feel free to do so in this class.
- Be respectful of your fellow students. While working together to build this community, we ask all members to:
- Share their unique experiences, values, and beliefs, if comfortable doing so.
- Listen deeply to one another.
- Honor the uniqueness of their peers.
- Create a respectful environment in this course and across the campus community.
- Check announcements or e-mails regularly. Announcements will serve as courtesy reminders and also point you to any new materials or changes.
- Do the pre-class assignments. In order to make the most of our class time, it is important that all students complete the pre-class assignments so we can jump into our application activities.
If you are unable to attend class, please contact the instructor as soon as possible via email at svenbuerki@boisestate.edu.
14 Expectations for Me
In support of my goal that every student be successful in this course, you can expect that I:
- Will be available to answer questions throughout the course. I encourage you to visit me during Student Hours to ask any questions about the course material or to further your curiosity in the subject matter. You may also email me with questions or concerns or ask to meet via Zoom or on campus outside of the Student Hours, and I will do my best to accommodate your needs.
- Will provide regular feedback on your work in a timely fashion. I will return completed work to you, with feedback, within one week of the due date.
- Will continuously work on improving your learning experience through my own class observations and based on your feedback. You will have the opportunity to provide feedback to me at the midpoint of the semester, and via the end of course evaluations.
15 This Class Welcomes Everyone
Students in this class represent a rich variety of backgrounds and perspectives. The (program/dept) is committed to providing an environment where similarities and differences are respected, supported, and valued. While working together to build this community, we ask all members to:
- share their unique experiences, values, and beliefs, if comfortable doing so.
- listen deeply to one another.
- honor the uniqueness of their peers.
- appreciate the opportunity we have to learn from each other in this community.
- use this opportunity together to discuss ways in which we can create a welcoming and respectful environment in this course and across the campus community.
- recognize opportunities to invite a community member to exhibit more respectful speech or behavior—and then also invite them into further conversation. We also expect community members to respond with gratitude and to take a moment of reflection when they receive such an invitation, rather than react immediately from defensiveness.
- keep confidential any discussions that the community has of a personal (or professional) nature, unless the speaker has given explicit permission to share what they have said.
As your instructor, my goal is to make sure that our learning environment is effective for everyone. This means, in part, that each student is encouraged to share perspectives relevant to the course material and that our class activities and discussions are conducted in a way that supports everyone’s learning.
16 Student Well-being
If you are struggling for any reason (e.g., family emergency, financial/basic needs, mental/physical health concerns, caregiving responsibilities, etc.) and believe these struggles may impact your performance in the course, I encourage you to reach out to me if you are comfortable doing so, and I will refer you to an appropriate university resource. You may also reach out directly to the outreach team in the Office of the Dean of Students at (208) 426-1527 or email studentoutreach@boisestate.edu for support. The Student Life Essentials page is also a great place to find helpful resources. If you notice a significant change in your mood, sleep, feelings of hopelessness or a lack of self worth, consider connecting immediately with Counseling Services (1529 Belmont Street, Norco Building) at (208) 426-1459 or email healthservices@boisestate.edu.
17 What Do you Need?
The university has many resources designed to support you as a learner and human being. Among these are:
- Albertsons Library provides a treasure trove of physical and electronic resources.
- As you enter the library, straight ahead you’ll find the Reference Desk, where librarians can help you find the information and resources you need.
- The Circulation Desk lets students borrow various technologies.
- The MakerLab on the second floor offers tools for student use, and there are friendly staff in the MakerLab to help you learn how.
- The Writing Center offers individual consultations tailored to your needs, including making sense of writing assignment instructions, brainstorming, crafting a thesis, organizing an essay, revisions, citations, and more.
- Counseling Services helps you tap into your strengths and find resources to deal more effectively with concerns that impact your pursuit of personal and academic goals. It emphasizes prevention and early detection and provides a broad spectrum of short-term counseling, consultative, evaluative, teaching, and training functions. Counseling staff consists of licensed counselors, psychologists, and closely supervised trainees/post-graduate interns.
- Food assistance: If you are hungry and cannot afford to purchase food, the campus has some resources to help you. You can visit the campus food pantry or get free meals in the campus dining hall.
18 Course Policies
18.1 Academic Integrity
Academic Excellence is a Shared Value at Boise State, and part of your responsibility in pursuing academic excellence includes avoiding cheating, plagiarism, and any other kind of academic misconduct. If I find a student responsible for academic misconduct in our class, the outcome of their choice to not fully engage in their learning might range from a ‘revise & resubmit’ up to an ‘F (failure) for the course.’ For more info, please read The Student Code of Conduct (Policy 2020), Section 7: Academic Misconduct Complaints, Violations, Processes and Sanctions.
18.2 Artifical Intelligence (AI) Use in The Course
In this course, I want to see your thoughts, understand your reasoning, and hear your voice. However, there are moments in this course where you might find it useful to use generative AI tools in support of your learning.
You may use generative AI tools for specified activities and assignments if their use supports, rather than undermines, your learning. While generative AI can help to advance your learning, its usefulness depends on the purpose of each activity or assignment. You will find guidelines for generative AI use in the instructions for each assignment; please read them very carefully, as these guidelines differ by assignment.
If you use ChatGPT, Gemini, Grammarly, Midjourney, or other AI tools in support of your work in this course, cite any ideas, text, images, or other media generated by the tool using the instructions and format of the Modern Language Association (MLA), American Psychological Association (APA), Chicago Manual of Style, or other citation style as appropriate. When you use a tool in an assignment, include a brief, clear description of how you used it. If you use generative AI, you must not let this tool replace your thinking and work. In fact, it is your responsibility to ensure you are fully engaging in learning and submitting authentic work. To learn more about how to learn successfully and avoid academic misconduct behaviors, please review the Student Code of Conduct with special attention to Section 8: Procedures for Academic Misconduct.
If you are unsure of whether or when to use generative AI tools in this course, please reach out to me. I am eager to learn about how we might use them in new ways to meaningfully advance your learning and prepare you for your future beyond Boise State.
18.3 Communicable Disease Policy
Boise State has a Communicable Disease Policy (Policy 9270) that guides everyone working and learning in our community. The policy has two key implications:
- Any illness covered by the policy must be reported to Boise State Public Health by the impacted individual,
- Faculty are required to accommodate any student impacted by any illness covered by the policy as directed by Boise State Public Health.
18.4 Late Work Policy
Due dates for all assignments are provided on the course schedule. Unless otherwise stated, assignments are due on those dates.
The instructor recognizes that unforeseen circumstances may arise. If this happens, please contact the instructor ahead of the due date, or as soon as possible afterwards, so that we can establish a plan for submitting the assignment as close to the original due date as possible.
Automatic Late Penalty:
- If a student fails to submit an assignment by the due date without a valid excuse, a 10% penalty will be applied to the assignment grade.
- In such cases, the instructor will reach out to the student to establish a new submission deadline and clarify expectations for completing the assignment.
Assignments that are not submitted within two weeks of the due date and for which the student has not communicated with the instructor will be recorded as Incomplete in Canvas until a plan is agreed upon.
19 Note on Course Content and Idaho Law
Under Idaho law (Section § 67-5909D), some university courses with content related to diversity, equity, inclusion, or critical theory may be subject to certain restrictions. However, the law affirms and does not limit free discussion in the learning environment. Like all Boise State courses, this course supports open inquiry, intellectual honesty, and respectful engagement with a range of perspectives, all of which are consistent with student rights and responsibilities described in the Student Code of Conduct (Policy 2020).
This course may include content that touches on concepts related to diversity, equity, inclusion (DEI), or critical theory—such as systemic inequality, cultural identity, or gender and race in society. If these topics are included, it is because they are relevant to the learning outcomes for this course and are explored to support critical thinking, deeper understanding, and respectful engagement with different perspectives. As part of the course, you may be asked to apply or explain ideas that come from a particular perspective. However, you are not required to adopt such perspectives as your own.
Our learning environment is a space for open dialogue and thoughtful discussion, including complex or challenging topics. Everyone is expected to engage with curiosity, listen respectfully, and contribute in ways that support a productive and welcoming learning environment. Boise State and the Idaho State Board of Education affirm the importance of free expression and academic inquiry. As outlined in SBOE Policy III.B:
“Membership in the academic community imposes on administrators, faculty members, other institutional employees, and students an obligation to respect the dignity of others, to acknowledge the right of others to express differing opinions, and to foster and defend intellectual honesty, freedom of inquiry and instruction, and free expression on and off the campus of an institution.”
Disruptive behavior that interferes with the learning environment will not be tolerated and may result in removal from this course, in line with university policy (See Policy 3240 Maintaining Effective Learning Environments).
In this course, I will foster critical discussion and analysis, and a respectful consideration of a wide range of ideas, in accordance with the Faculty Code of Rights, Responsibilities, and Conduct (Policy 4000). You are encouraged to think critically, question ideas, and form your own conclusions. As always, you have the freedom to choose courses that align with your academic goals—if you have concerns about course content, please talk with your instructor or advisor. Refer to the academic calendar for important deadlines related to course withdrawal.
To learn more about the law and its impact at Boise State, visit the Provost Office’s Information Regarding Section 67-5909D page.
20 References
21 Appendix 1
Citations of all R packages used to generate this report.
[1] J. Allaire, Y. Xie, C. Dervieux, et al. rmarkdown: Dynamic Documents for R. R package version 2.30. 2025. https://github.com/rstudio/rmarkdown.
[2] C. Boettiger. knitcitations: Citations for Knitr Markdown Files. R package version 1.0.12. 2021. https://github.com/cboettig/knitcitations.
[3] M. C. Koohafkan. kfigr: Integrated Code Chunk Anchoring and Referencing for R Markdown Documents. R package version 1.2.1. 2021. https://github.com/mkoohafkan/kfigr.
[4] R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Vienna, Austria, 2025. https://www.R-project.org/.
[5] H. Wickham, J. Bryan, M. Barrett, et al. usethis: Automate Package and Project Setup. R package version 3.2.1. 2025. https://usethis.r-lib.org.
[6] H. Wickham, R. François, L. Henry, et al. dplyr: A Grammar of Data Manipulation. R package version 1.1.4. 2023. https://dplyr.tidyverse.org.
[7] H. Wickham, J. Hester, W. Chang, et al. devtools: Tools to Make Developing R Packages Easier. R package version 2.4.6. 2025. https://devtools.r-lib.org/.
[8] Y. Xie. bookdown: Authoring Books and Technical Documents with R Markdown. Boca Raton, Florida: Chapman and Hall/CRC, 2016. ISBN: 978-1138700109. https://bookdown.org/yihui/bookdown.
[9] Y. Xie. bookdown: Authoring Books and Technical Documents with R Markdown. R package version 0.46. 2025. https://github.com/rstudio/bookdown.
[10] Y. Xie. Dynamic Documents with R and knitr. 2nd. ISBN 978-1498716963. Boca Raton, Florida: Chapman and Hall/CRC, 2015. https://yihui.org/knitr/.
[11] Y. Xie. formatR: Format R Code Automatically. R package version 1.14. 2023. https://github.com/yihui/formatR.
[12] Y. Xie. “knitr: A Comprehensive Tool for Reproducible Research in R”. In: Implementing Reproducible Computational Research. Ed. by V. Stodden, F. Leisch and R. D. Peng. ISBN 978-1466561595. Chapman and Hall/CRC, 2014.
[13] Y. Xie. knitr: A General-Purpose Package for Dynamic Report Generation in R. R package version 1.50. 2025. https://yihui.org/knitr/.
[14] Y. Xie and J. Allaire. tufte: Tufte’s Styles for R Markdown Documents. R package version 0.14.0. 2025. https://github.com/rstudio/tufte.
[15] Y. Xie, J. Allaire, and G. Grolemund. R Markdown: The Definitive Guide. Boca Raton, Florida: Chapman and Hall/CRC, 2018. ISBN: 9781138359338. https://bookdown.org/yihui/rmarkdown.
[16] Y. Xie, C. Dervieux, and E. Riederer. R Markdown Cookbook. Boca Raton, Florida: Chapman and Hall/CRC, 2020. ISBN: 9780367563837. https://bookdown.org/yihui/rmarkdown-cookbook.
[17] H. Zhu. kableExtra: Construct Complex Table with kable and Pipe Syntax. R package version 1.4.0. 2024. http://haozhu233.github.io/kableExtra/.