Sagebrush Genome Project
1 Aim
The Sagebrush Genome Project supports the overarching aim of the GEM3 project, which is to understand genome to phenome (G2P) mechanisms that translate to adaptive capacity of sagebrush (Artemisia tridentata) populations. More specifically, in this project, we will be focusing on assessing sagebrush adaptive capacity to respond to drought and heat stress caused by climate change.
2 Objectives
This project is subdivided into 5 objectives as follows:
- Individual lines: Developing and applying an in vitro tissue culture approach to propagate sagebrush individual lines supporting objectives 2-4. This objective is key since preliminary analyses demonstrated that the sagebrush genome is highly out-bred. For these reasons, we are aiming at not only produce enough biomass of the same individual line to produce the reference genome, but also maintain individual lines to support genotype-by-environment (GxE) experiments (objective 4).
- Reference genome: Sequencing, phasing and annotating the diploid genome of basin big sagebrush (Artemisia tridentata subsp. tridentata). This taxon as 2n=2x=18 chromosomes and a 2C genome size of ca. 9.5Gbp (see Barron et al., 2020). To deliver such complex genome, we aim at generating 50-100X raw data coverage (haploid sagebrush genome size: 4.5 Gbp) using PacBio technology and phase it using proximity ligation data. Such wet-lab procedures are requiring the production of ca. 120 gr. of leaf biomass. Annotation of the reference genome will be conducted based on RNA-seq data generated during objective 4 (through GxE experiments). This will allow focusing on annotating genes involved in responding to drought and heat stresses and therefore assess G2P mechanisms.
- Comparative genomics: To further investigate genomic differences between sagebrush populations, a re-sequencing approach coupled with pan-genomic analyses will be applied to compare genomes from individual lines growing in contrasting climates (in terms of drought and heat stresses).
- Common gardens: GxE experiments focusing on measuring the phenotypic and transcriptomic responses of sagebrush individual lines to drought, heat and drought+heat stresses will be conducted. Using clones will allow to disentangle respective contributions of genomic vs. phenotypic plasticty processes on the adaptive capacity of sagebrush genotypes. An overarching objective here is to identify adaptive loci key in drought, heat and drought+heat responses. These adaptive loci will then be used to model sagebrush adaptive capacity at population level in objective 5.
- Landscape genomics: Apply adaptive loci (sequenced using a baiting approach) on sagebrush individuals collected from populations representing climatic and geographic range of the species. First, field collections will be biobanked (= conducting DNA extractions and establishing a seed bank) and herbarium specimens digitized. The age of each collected individual will be estimated and compared to onset of megadrought in the US (Williams et al., 2020). This preliminary data will provide hypotheses on the establishment of individuals and populations in the landscape and the potential effects of drought on the gene pool. We hypothesize that the effect of megadrought on established individuals will be less dramatic than on those establishing post megadrought. This means that if sagebrush megadrought (and associated heat) response is genetically driven then more genetic diversity (based on adaptive loci inferred in objective 4) will be observed for individuals established prior to megadrought, whereas those established post megadrought will show a strong genetic bottleneck effect corresponding to natural selection processes acting upon sageberush populations.
3 Timetable
The timetable associated to our objectives is displayed in Figure 3.1.
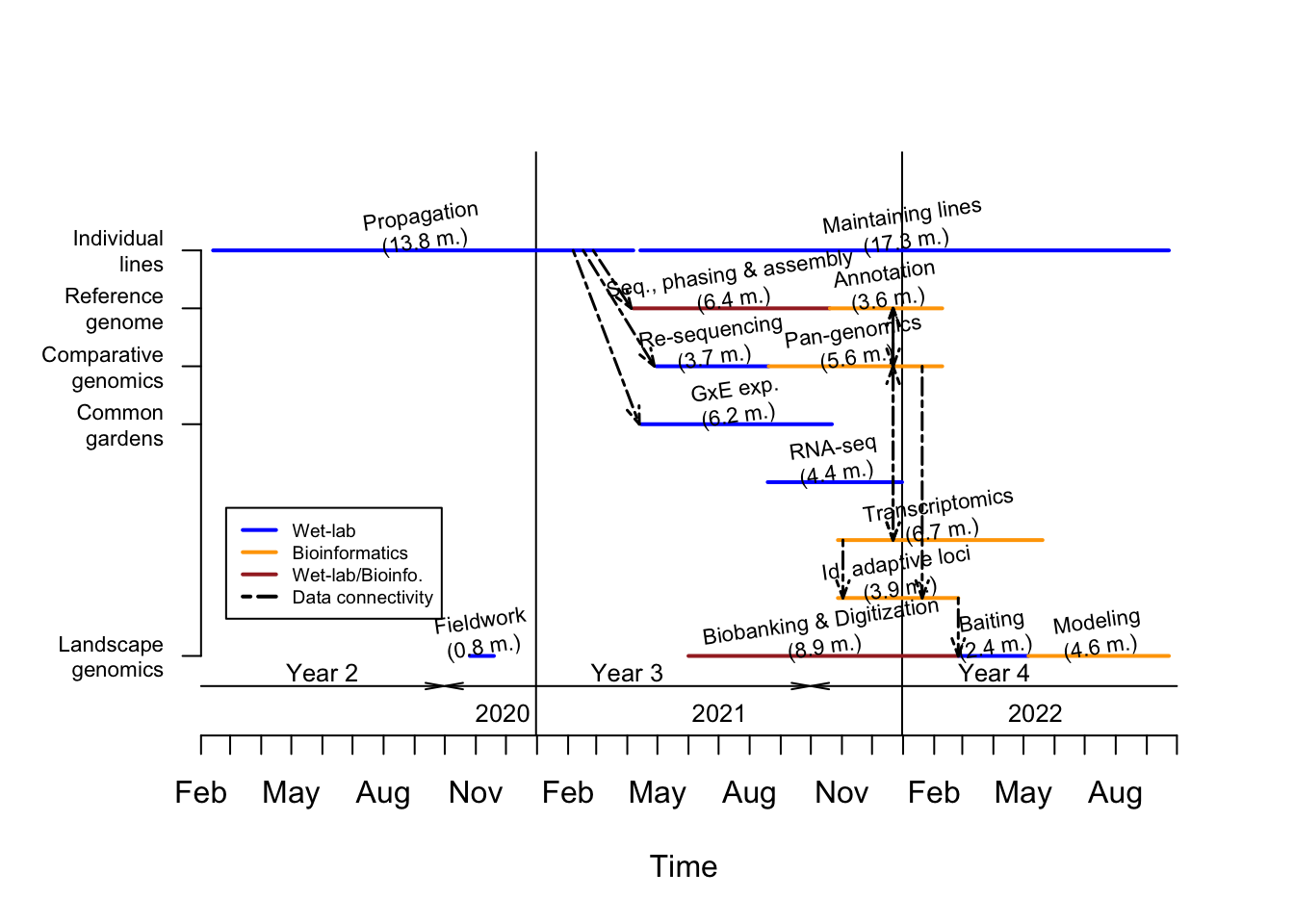
Figure 3.1: Timetable associated to production of data for the sagebrush genome project. Please see text for more details.
4 Project structure
Each of the objective described above and in Figure 3.1 are further detailed in the following webpage:
- Propagation program: This webpage covers the tissue culture methods applied in this project together with data on individual lines maintained at BSU.
- Sequencing: This webpage describes the approach used to sequence, phase and annotate sagebrush genome.
- Annotation: This webpage provides insights into the GxE experiments used to estimate the effect of drought, heat and drought+heat on sagebrush individual lines. RNA-seq data produced by these experiments will be used to annotate sagebrush genome and identify adaptive loci.
References
Barron, R., P. Martinez, M. Serpe, and S. Buerki. 2020. Development of an in vitro method of propagation for artemisia tridentata subsp. Tridentata to support genome sequencing and genotype-by-environment research. Plants 9: Available at: https://www.mdpi.com/2223-7747/9/12/1717.
Williams, A.P., E.R. Cook, J.E. Smerdon, B.I. Cook, J.T. Abatzoglou, K. Bolles, S.H. Baek, et al. 2020. Large contribution from anthropogenic warming to an emerging north american megadrought. Science 368: 314–318.